This is built with spread2() and is still experimental.
This one differs from other attempts in that it treats the advection and
dispersal as mathematical vectors that are added together.
They are "rounded" to pixel centres.
spread3(
start,
rasQuality,
rasAbundance,
advectionDir,
advectionMag,
meanDist,
dispersalKernel = "exponential",
sdDist = 1,
plot.it = 2,
minNumAgents = 50,
verbose = getOption("LandR.verbose", 0),
saveStack = NULL,
skipChecks = FALSE
)Arguments
- start
Raster indices from which to initiate dispersal
- rasQuality
A raster with habitat quality. Currently, must be scaled from 0 to 1, i.e., a probability of "settling"
- rasAbundance
A raster where each pixel represents the number of "agents" or pseudo-agents contained. This number of agents, will be spread horizontally, and distributed from each pixel that contains a non-zero non NA value.
- advectionDir
A single number or
RasterLayerin degrees from North = 0 (though it will use radians if all values areabs(advectionDir) > 2 * pi). This indicates the direction of advective forcing (i.e., wind).- advectionMag
A single number or
RasterLayerin distance units of therasQuality, e.g., meters, indicating the relative forcing that will occur. It is imposed on the total event, i.e., if themeanDistis10000, andadvectionMagis5000, then the expected distance (i.e.,63%of agents) will have settled by15000map units.- meanDist
A single number indicating the mean distance parameter in map units (not pixels), for a negative exponential distribution dispersal kernel (e.g.,
dexp). This will mean that63%of agents will have settled at thismeanDist(still experimental).- dispersalKernel
One of either
"exponential"or"weibull".- sdDist
A single number indicating the
sdparameter of a two-parameterdispersalKernel. Defaults to1, which is the same as theexponentialdistribution.- plot.it
Numeric. With increasing numbers above 0, there will be plots produced during iterations. Currently, only 0, 1, or 2+ are distinct.
- minNumAgents
Single numeric indicating the minimum number of agents to consider all dispersing finished. Default is 50.
- verbose
Numeric. With increasing numbers above 0, there will be more messages produced. Currently, only 0, 1, or 2+ are distinct.
- saveStack
If provided as a character string, it will save each iteration as part of a
rasterStackto disk upon exit.- skipChecks
Logical. If
TRUE, assertions will be skipped (faster, but could miss problems)
Value
A data.table with all information used during the spreading
Examples
## these tests are fairly heavy, so don't run during automated tests
#########################################################
# Simple case, no variation in rasQuality, numeric advectionDir and advectionMag
#########################################################
# \donttest{
library(terra)
origDTThreads <- data.table::setDTthreads(2L)
origNcpus <- options(Ncpus = 2L)
maxDim <- 10000
ras <- terra::rast(terra::ext(c(0, maxDim, 0, maxDim)), res = 100, vals = 0)
rasQuality <- terra::rast(ras)
rasQuality[] <- 1
rasAbundance <- terra::rast(rasQuality)
rasAbundance[] <- 0
# startPixel <- middlePixel(rasAbundance)
startPixel <- sample(seq(terra::ncell(rasAbundance)), 30)
rasAbundance[startPixel] <- 1000
advectionDir <- 70
advectionMag <- 4 * res(rasAbundance)[1]
meanDist <- 2600
# Test the dispersal kernel -- create a function
plotDispersalKernel <- function(out, meanAdvectionMag) {
out[, disGroup := round(distance / 100) * 100]
freqs <- out[, .N, by = "disGroup"]
freqs[, `:=`(cumSum = cumsum(N), N = N)]
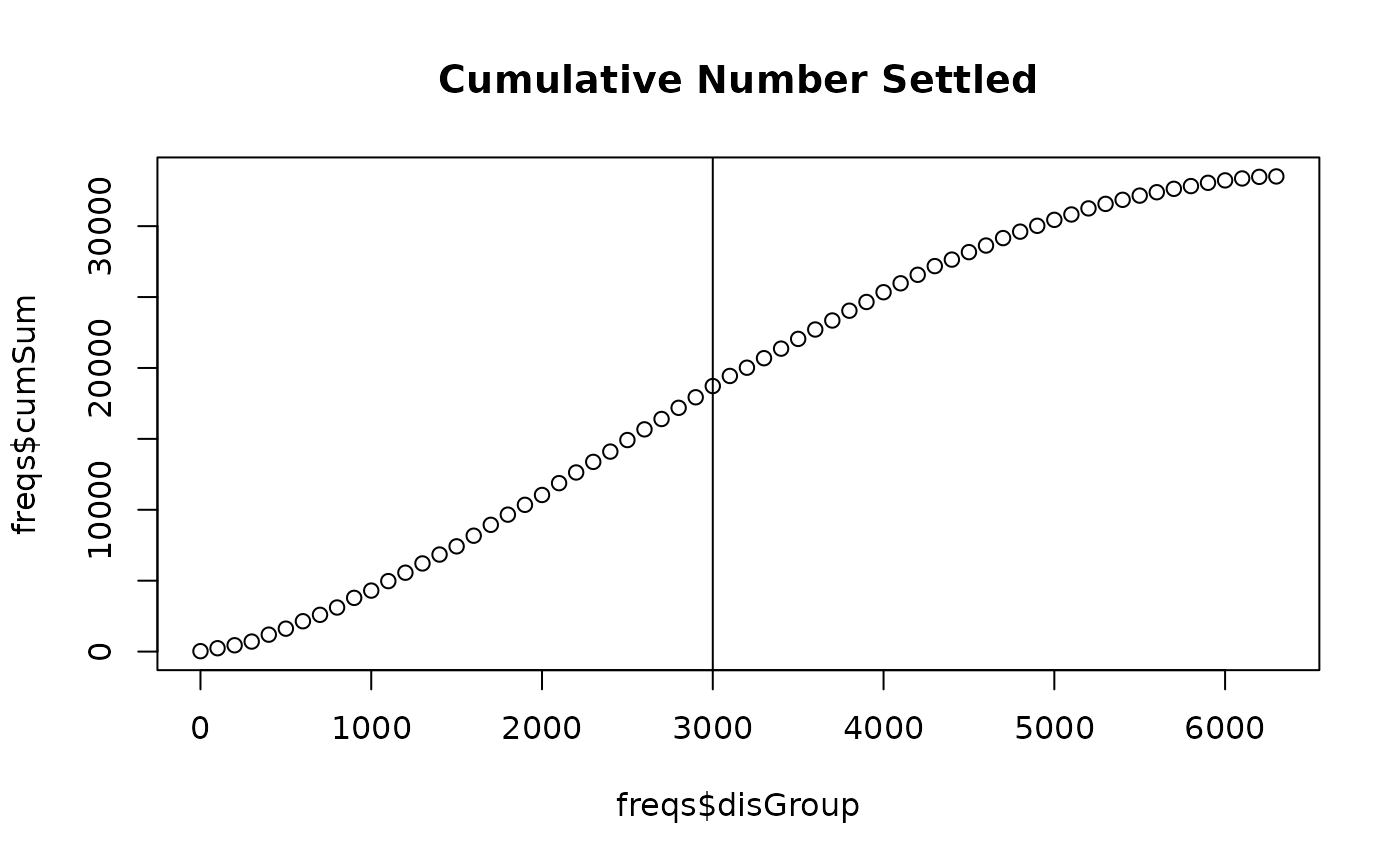
plot(freqs$disGroup, freqs$cumSum, # addTo = "CumulativeNumberSettled",
main = "Cumulative Number Settled") # can plot the distance X number
abline(v = meanAdvectionMag + meanDist)
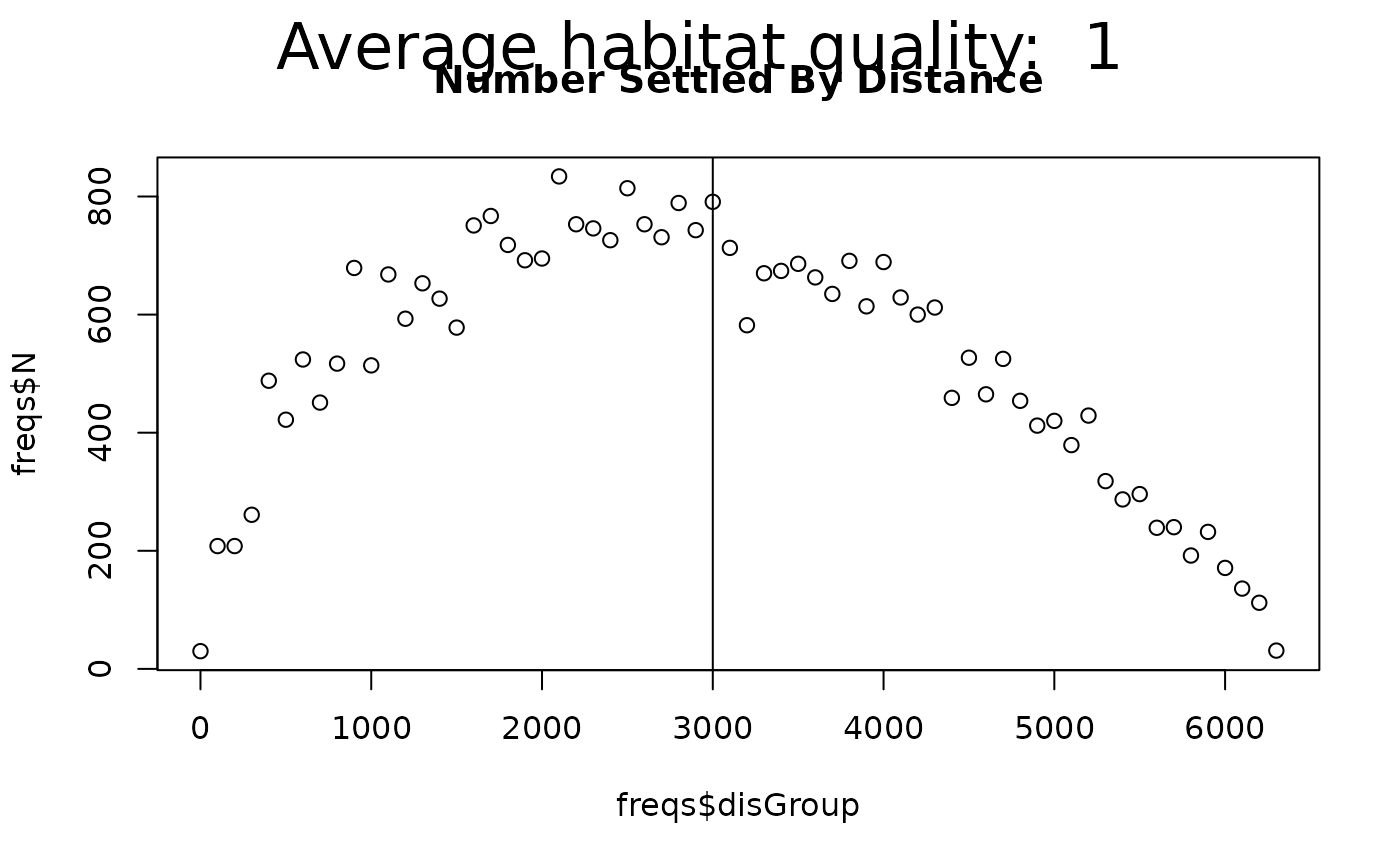
newTitle <- "Number Settled By Distance"
plot(freqs$disGroup, freqs$N, # addTo = gsub(" ", "", newTitle),
main = newTitle) # can plot the distance X number
abline(v = meanAdvectionMag + meanDist)
# should be 0.63:
freqs[disGroup == meanAdvectionMag + meanDist, cumSum] / tail(freqs, 1)[, cumSum]
mtext(side = 3, paste("Average habitat quality: ",
round(mean(rasQuality[], na.rm = TRUE), 2)),
outer = TRUE, line = -2, cex = 2)
}
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = meanDist, verbose = 2,
plot.it = interactive())
#> assuming that advectionDir is in geographic degrees(i.e., North is 0)
#> Iteration 1
#> Number still dispersing 29942.6974241127
#> Iteration 2
#> Number still dispersing 29845.2355120556
#> Iteration 3
#> Number still dispersing 29684.8907058857
#> Iteration 4
#> Number still dispersing 28961.0363094562
#> Iteration 5
#> Number still dispersing 28689.172882837
#> Iteration 6
#> Number still dispersing 28297.5381929262
#> Iteration 7
#> Number still dispersing 27874.7434601112
#> Iteration 8
#> Number still dispersing 26484.7181518992
#> Iteration 9
#> Number still dispersing 25582.8367937818
#> Iteration 10
#> Number still dispersing 24950.0287991057
#> Iteration 11
#> Number still dispersing 23810.3470626647
#> Iteration 12
#> Number still dispersing 23138.6878916629
#> Iteration 13
#> Number still dispersing 22478.8630156612
#> Iteration 14
#> Number still dispersing 21493.294194617
#> Iteration 15
#> Number still dispersing 20878.3755159988
#> Iteration 16
#> Number still dispersing 20230.8132430021
#> Iteration 17
#> Number still dispersing 19558.2859208612
#> Iteration 18
#> Number still dispersing 18798.6791665058
#> Iteration 19
#> Number still dispersing 18114.4685714799
#> Iteration 20
#> Number still dispersing 17335.5307185944
#> Iteration 21
#> Number still dispersing 16617.7831503639
#> Iteration 22
#> Number still dispersing 15886.9462151914
#> Iteration 23
#> Number still dispersing 15193.9920401682
#> Iteration 24
#> Number still dispersing 14475.3920643484
#> Iteration 25
#> Number still dispersing 13773.1118090256
#> Iteration 26
#> Number still dispersing 13098.0798618491
#> Iteration 27
#> Number still dispersing 12267.94522178
#> Iteration 28
#> Number still dispersing 11605.0397688509
#> Iteration 29
#> Number still dispersing 10952.5137443267
#> Iteration 30
#> Number still dispersing 9991.87560873532
#> Iteration 31
#> Number still dispersing 9320.07536604412
#> Iteration 32
#> Number still dispersing 8608.22893176744
#> Iteration 33
#> Number still dispersing 8019.3137539911
#> Iteration 34
#> Number still dispersing 7419.6941039023
#> Iteration 35
#> Number still dispersing 6824.49671291095
#> Iteration 36
#> Number still dispersing 6317.9586758525
#> Iteration 37
#> Number still dispersing 5865.93380825119
#> Iteration 38
#> Number still dispersing 5377.47629354724
#> Iteration 39
#> Number still dispersing 4919.23529026206
#> Iteration 40
#> Number still dispersing 4529.38133634955
#> Iteration 41
#> Number still dispersing 4118.66865850671
#> Iteration 42
#> Number still dispersing 3713.52728051165
#> Iteration 43
#> Number still dispersing 3336.93962976748
#> Iteration 44
#> Number still dispersing 2980.35984067639
#> Iteration 45
#> Number still dispersing 2697.3979402356
#> Iteration 46
#> Number still dispersing 2415.25223056504
#> Iteration 47
#> Number still dispersing 2116.82733879199
#> Iteration 48
#> Number still dispersing 1918.09641472157
#> Iteration 49
#> Number still dispersing 1647.41511007985
#> Iteration 50
#> Number still dispersing 1442.02030855928
#> Iteration 51
#> Number still dispersing 1252.53030232797
#> Iteration 52
#> Number still dispersing 1068.17138783792
#> Iteration 53
#> Number still dispersing 898.580870314861
#> Iteration 54
#> Number still dispersing 737.905987640094
#> Iteration 55
#> Number still dispersing 656.285745925141
#> Iteration 56
#> Number still dispersing 514.584219321668
#> Iteration 57
#> Number still dispersing 440.522016969407
#> Iteration 58
#> Number still dispersing 388.674735204424
#> Iteration 59
#> Number still dispersing 295.769875692062
#> Iteration 60
#> Number still dispersing 215.171705314287
#> Iteration 61
#> Number still dispersing 178.493658565594
#> Iteration 62
#> Number still dispersing 97.6754223744511
#> Iteration 63
#> Number still dispersing 21.8713131945417
plotDispersalKernel(out, advectionMag)
 # The next examples are potentially time consuming; avoid on automated testing
if (interactive()) {
#########################################################
### The case of variable quality raster
#########################################################
rasQuality <- terra::rast(system.file("extdata", "rasQuality.tif",
package = "SpaDES.tools"))
terra::crs(rasQuality) <- system.file("extdata", "targetCRS.rds", package = "SpaDES.tools") |>
readRDS() |>
slot("projargs")
mask <- rasQuality < 5
rasQuality[mask[] %in% TRUE] <- 0
# rescale so min is 0.75 and max is 1
rasQuality[] <- rasQuality[] / (reproducible::maxFn(rasQuality) * 4) + 1 / 4
rasAbundance <- terra::rast(rasQuality)
rasAbundance[] <- 0
startPixel <- sample(seq(ncell(rasAbundance)), 300)
rasAbundance[startPixel] <- 1000
advectionDir <- 75
advectionMag <- 4 * res(rasAbundance)[1]
meanDist <- 2600
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = meanDist, verbose = 2,
plot.it = interactive())
if (interactive()) {
plotDispersalKernel(out, advectionMag)
}
###############################################################################
### The case of variable quality raster, raster for advectionDir & advectionMag
###############################################################################
maxDim <- 10000
ras <- terra::rast(terra::ext(c(0, maxDim, 0, maxDim)), res = 100, vals = 0)
rasQuality <- terra::rast(ras)
rasQuality[] <- 1
rasAbundance <- terra::rast(rasQuality)
rasAbundance[] <- NA
# startPixel <- middlePixel(rasAbundance)
startPixel <- sample(seq(ncell(rasAbundance)), 25)
rasAbundance[startPixel] <- 1000
# raster for advectionDir
advectionDir <- terra::rast(system.file("extdata", "advectionDir.tif",
package = "SpaDES.tools"))
crs(advectionDir) <- crs(rasQuality)
# rescale so min is 0.75 and max is 1
advectionDir[] <- advectionDir[] / (reproducible::maxFn(advectionDir)) * 180
# raster for advectionMag
advectionMag <- terra::rast(system.file("extdata", "advectionMag.tif",
package = "SpaDES.tools"))
crs(advectionMag) <- crs(rasQuality)
# rescale so min is 0.75 and max is 1
advectionMag[] <- advectionMag[] / (reproducible::maxFn(advectionMag)) * 600
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = meanDist, verbose = 2,
plot.it = interactive())
if (interactive()) {
names(advectionDir) <- "Wind direction"
names(advectionMag) <- "Wind speed"
names(rasAbundance) <- "Initial abundances"
terra::plot(c(advectionDir, advectionMag, rasAbundance))
plotDispersalKernel(out, mean(advectionMag[]))
}
#########################################
# save iterations to a stack to make animated GIF
########################################
tmpStack <- tempfile(pattern = "stackToAnimate", fileext = ".tif")
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = 2600, verbose = 2,
plot.it = interactive(), saveStack = tmpStack)
## This animates the series of images into an animated GIF
if (require(animation, quietly = TRUE)) {
out2 <- terra::rast(tmpStack)
gifName <- file.path(tempdir(), "animation.gif")
# Only works on some systems; may need to configure
# Works on Windows without system adjustments
if (identical(.Platform$OS.type, "windows"))
saveGIF(interval = 0.1, movie.name = gifName, expr = {
for (i in seq(length(names(out2)))) terra::plot(out2[[i]])
})
}
}
# clean up
data.table::setDTthreads(origDTThreads)
options(Ncpus = origNcpus)
# }
# The next examples are potentially time consuming; avoid on automated testing
if (interactive()) {
#########################################################
### The case of variable quality raster
#########################################################
rasQuality <- terra::rast(system.file("extdata", "rasQuality.tif",
package = "SpaDES.tools"))
terra::crs(rasQuality) <- system.file("extdata", "targetCRS.rds", package = "SpaDES.tools") |>
readRDS() |>
slot("projargs")
mask <- rasQuality < 5
rasQuality[mask[] %in% TRUE] <- 0
# rescale so min is 0.75 and max is 1
rasQuality[] <- rasQuality[] / (reproducible::maxFn(rasQuality) * 4) + 1 / 4
rasAbundance <- terra::rast(rasQuality)
rasAbundance[] <- 0
startPixel <- sample(seq(ncell(rasAbundance)), 300)
rasAbundance[startPixel] <- 1000
advectionDir <- 75
advectionMag <- 4 * res(rasAbundance)[1]
meanDist <- 2600
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = meanDist, verbose = 2,
plot.it = interactive())
if (interactive()) {
plotDispersalKernel(out, advectionMag)
}
###############################################################################
### The case of variable quality raster, raster for advectionDir & advectionMag
###############################################################################
maxDim <- 10000
ras <- terra::rast(terra::ext(c(0, maxDim, 0, maxDim)), res = 100, vals = 0)
rasQuality <- terra::rast(ras)
rasQuality[] <- 1
rasAbundance <- terra::rast(rasQuality)
rasAbundance[] <- NA
# startPixel <- middlePixel(rasAbundance)
startPixel <- sample(seq(ncell(rasAbundance)), 25)
rasAbundance[startPixel] <- 1000
# raster for advectionDir
advectionDir <- terra::rast(system.file("extdata", "advectionDir.tif",
package = "SpaDES.tools"))
crs(advectionDir) <- crs(rasQuality)
# rescale so min is 0.75 and max is 1
advectionDir[] <- advectionDir[] / (reproducible::maxFn(advectionDir)) * 180
# raster for advectionMag
advectionMag <- terra::rast(system.file("extdata", "advectionMag.tif",
package = "SpaDES.tools"))
crs(advectionMag) <- crs(rasQuality)
# rescale so min is 0.75 and max is 1
advectionMag[] <- advectionMag[] / (reproducible::maxFn(advectionMag)) * 600
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = meanDist, verbose = 2,
plot.it = interactive())
if (interactive()) {
names(advectionDir) <- "Wind direction"
names(advectionMag) <- "Wind speed"
names(rasAbundance) <- "Initial abundances"
terra::plot(c(advectionDir, advectionMag, rasAbundance))
plotDispersalKernel(out, mean(advectionMag[]))
}
#########################################
# save iterations to a stack to make animated GIF
########################################
tmpStack <- tempfile(pattern = "stackToAnimate", fileext = ".tif")
out <- spread3(rasAbundance = rasAbundance,
rasQuality = rasQuality,
advectionDir = advectionDir,
advectionMag = advectionMag,
meanDist = 2600, verbose = 2,
plot.it = interactive(), saveStack = tmpStack)
## This animates the series of images into an animated GIF
if (require(animation, quietly = TRUE)) {
out2 <- terra::rast(tmpStack)
gifName <- file.path(tempdir(), "animation.gif")
# Only works on some systems; may need to configure
# Works on Windows without system adjustments
if (identical(.Platform$OS.type, "windows"))
saveGIF(interval = 0.1, movie.name = gifName, expr = {
for (i in seq(length(names(out2)))) terra::plot(out2[[i]])
})
}
}
# clean up
data.table::setDTthreads(origDTThreads)
options(Ncpus = origNcpus)
# }